skelm.LargeELMRegressor¶
- class skelm.LargeELMRegressor(SLFNs: Iterable[SLFN], solver: Solver)[source]¶
ELM Regressor for larger-than-memory problems.
Uses Dask for batch analysis of data in Parquet files.
Attention
Why do I need Parquet files?
Parquet files provide necessary information about the data without loading whole file content from disk. It makes a tremendous runtime difference compared to simpler .csv or .json file formats. Reading from files saves memory by loading data in small chunks, supporting arbitrary large input files. It also solves current memory leaks with Numpy matrix inputs in Dask.
Any data format can be easily converted to Parquet, see Analytical methods section.
HDF5 is almost as good as Parquet, but performs worse with Dask due to internal data layout.
- fit(X, y=None, sync_every=10)[source]¶
Fits an ELM with data in a bunch of files.
Model will use the set of features from the first file. Same features must have same names across the whole dataset.
Does not support sparse data.
Original features and bias are added to the end of data, for easier rechunk-merge. This way full chunks of hidden neuron outputs stay intact.
- Parameters:
X ([str]) – List of input data files in Parquet format.
y ([str]) – List of target data files in Parquet format.
sync_every (int or None) – Synchronize computations after this many files are processed. None for running without synchronization. Less synchronization improves run speed with smaller data files, but may result in large swap space usage for large data problems. Use smaller number for more frequent synchronization if swap space becomes a problem.
- property n_neurons¶
Number of neurons in ELM model
- predict(X)[source]¶
Prediction works with both lists of Parquet files and numeric arrays.
- Parameters:
X (array-like, [str]) – Input data as list of Parquet files, or as a numeric array.
- Returns:
Yh – Predicted values for all input samples.
Attention
Returns all outputs as a single in-memory array!
Danger of running out out memory for high-dimensional outputs, if a large set of input files is provided. Feed data in smaller batches in such case.
- Return type:
array, shape (n_samples, n_outputs)
- score(X, y, sample_weight=None)¶
Return the coefficient of determination of the prediction.
The coefficient of determination
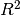 is defined as
is defined as
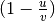 , where
, where 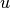 is the residual
sum of squares
is the residual
sum of squares ((y_true - y_pred)** 2).sum()and is the total sum of squares
is the total sum of squares ((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a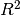 score of 0.0.
score of 0.0.- Parameters:
X (array-like of shape (n_samples, n_features)) – Test samples. For some estimators this may be a precomputed kernel matrix or a list of generic objects instead with shape
(n_samples, n_samples_fitted), wheren_samples_fittedis the number of samples used in the fitting for the estimator.y (array-like of shape (n_samples,) or (n_samples, n_outputs)) – True values for X.
sample_weight (array-like of shape (n_samples,), default=None) – Sample weights.
- Returns:
score –
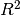 of
of self.predict(X)w.r.t. y.- Return type:
Notes
The
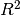 score used when calling
score used when calling scoreon a regressor usesmultioutput='uniform_average'from version 0.23 to keep consistent with default value ofr2_score(). This influences thescoremethod of all the multioutput regressors (except forMultiOutputRegressor).