skelm.ELMRegressor¶
- class skelm.ELMRegressor(alpha=1e-07, batch_size=None, include_original_features=False, n_neurons=None, ufunc='tanh', density=None, pairwise_metric=None, random_state=None)[source]¶
Extreme Learning Machine for regression problems.
This model solves a regression problem, that is a problem of predicting continuous outputs. It supports multi-variate regression (when
yis a 2d array of shape [n_samples, n_targets].) ELM usesL2regularization, and optionally includes the original data features to capture linear dependencies in the data natively.- Parameters:
alpha (float) –
Regularization strength; must be a positive float. Larger values specify stronger effect. Regularization improves model stability and reduces over-fitting at the cost of some learning capacity. The same value is used for all targets in multi-variate regression.
The optimal regularization strength is suggested to select from a large range of logarithmically distributed values, e.g.
![[10^{-5}, 10^{-4}, 10^{-3}, ..., 10^4, 10^5]](../_images/math/f81aed5cd381c497c0d2ddf5c4406d46277d53c0.png) . A small default
regularization value of
. A small default
regularization value of 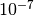 should always be present to counter numerical instabilities
in the solution; it does not affect overall model performance.
should always be present to counter numerical instabilities
in the solution; it does not affect overall model performance.Attention
The model may automatically increase the regularization value if the solution becomes unfeasible otherwise. The actual used value contains in
alpha_attribute.batch_size (int, optional) – Actual computations will proceed in batches of this size, except the last batch that may be smaller. Default behavior is to process all data at once with <10,000 samples, otherwise use batches of size 2000.
include_original_features (boolean, default=False) – Adds extra hidden layer neurons that simpy copy the input data features, adding a linear part to the final model solution that can directly capture linear relations between data and outputs. Effectively increases n_neurons by n_inputs leading to a larger model. Including original features is generally a good thing if the number of data features is low.
n_neurons (int or [int], optional) –
Number of hidden layer neurons in ELM model, controls model size and learning capacity. Generally number of neurons should be less than the number of training data samples, as otherwise the model will learn the training set perfectly resulting in overfitting.
Several different kinds of neurons can be used in the same model by specifying a list of neuron counts. ELM will create a separate neuron type for each element in the list. In that case, the following attributes
ufunc,densityandpairwise_metricshould be lists of the same length; default values will be automatically expanded into a list.Note
Models with <1,000 neurons are very fast to compute, while GPU acceleration is efficient starting from 1,000-2,000 neurons. A standard computer should handle up to 10,000 neurons. Very large models will not fit in memory but can still be trained by an out-of-core solver.
ufunc ({'tanh', 'sigm', 'relu', 'lin' or callable}, or a list of those (see n_neurons)) –
- Transformation function of hidden layer neurons. Includes the following options:
’tanh’ for hyperbolic tangent
’sigm’ for sigmoid
’relu’ for rectified linear unit (clamps negative values to zero)
’lin’ for linear neurons, transformation function does nothing
any custom callable function like members of
Numpu.ufunc
density (float in range (0, 1], or a list of those (see n_neurons), optional) – Specifying density replaces dense projection layer by a sparse one with the specified density of the connections. For instance,
density=0.1means each hidden neuron will be connected to a random 10% of input features. Useful for working on very high-dimensional data, or for large numbers of neurons.pairwise_metric ({'euclidean', 'cityblock', 'cosine' or other}, or a list of those (see n_neurons), optional) –
Specifying pairwise metric replaces multiplicative hidden neurons by distance-based hidden neurons. This ELM model is known as Radial Basis Function ELM (RBF-ELM).
Note
Pairwise function neurons ignore ufunc and density.
Typical metrics are euclidean, cityblock and cosine. For a full list of metrics check the webpage of
sklearn.metrics.pairwise_distances.random_state (int, RandomState instance or None, optional, default None) – The seed of the pseudo random number generator to use when generating random numbers e.g. for hidden neuron parameters. Random state instance is passed to lower level objects and routines. Use it for repeatable experiments.
- ufunc_¶
Tranformation function of hidden neurons.
- Type:
function
Examples
Combining ten sigmoid and twenty RBF neurons in one model:
>>> model = ELMRegressor(n_neurons=(10, 20), ... ufunc=('sigm', None), ... density=(None, None), ... pairwise_metric=(None, 'euclidean'))
Default values in multi-neuron ELM are automatically expanded to a list
>>> model = ELMRegressor(n_neurons=(10, 20), ... ufunc=('sigm', None), ... pairwise_metric=(None, 'euclidean'))
>>> model = ELMRegressor(n_neurons=(30, 30), ... pairwise_metric=('cityblock', 'cosine'))
- __init__(alpha=1e-07, batch_size=None, include_original_features=False, n_neurons=None, ufunc='tanh', density=None, pairwise_metric=None, random_state=None)¶
Scikit-ELM’s version of __init__, that only saves input parameters and does nothing else.
- fit(X, y) ScikitELM¶
Reset model and fit on the given data.
- Parameters:
X ({array-like, sparse matrix}, shape (n_samples, n_features)) – Training data samples.
y (array-like, shape (n_samples,) or (n_samples, n_outputs)) – Target values used as real numbers.
- Returns:
self – Returns self.
- Return type:
- get_params(deep=True)¶
Get parameters for this estimator.
- partial_fit(X, y=None, forget=False, compute_output_weights=True) ScikitELM¶
Update model with a new batch of data.
Output weight computation can be temporary turned off for faster processing. This will mark model as not fit. Enable compute_output_weights in the final call to partial_fit.
- Parameters:
X ({array-like, sparse matrix}, shape=[n_samples, n_features]) – Training input samples
y (array-like, shape=[n_samples, n_targets]) – Training targets
forget (boolean, default False) – Performs a negative update, effectively removing the information given by training samples from the model. Output weights need to be re-computed after forgetting data. Forgetting data that have not been learned before leads to unpredictable results.
compute_output_weights (boolean, optional, default True) –
Whether to compute new output weights (coef_, intercept_). Disable this in intermediate partial_fit steps to run computations faster, then enable in the last call to compute the new solution.
Note
Solution can be updated without extra data by setting X=None and y=None.
- Example:
>>> model.partial_fit(X_1, y_1) ... model.partial_fit(X_2, y_2) ... model.partial_fit(X_3, y_3)
- Faster:
>>> model.partial_fit(X_1, y_1, compute_output_weights=False) ... model.partial_fit(X_2, y_2, compute_output_weights=False) ... model.partial_fit(X_3, y_3)
- predict(X) _SupportsArray[dtype] | _NestedSequence[_SupportsArray[dtype]] | bool | int | float | complex | str | bytes | _NestedSequence[bool | int | float | complex | str | bytes]¶
Predict real valued outputs for new inputs X.
- Parameters:
X ({array-like, sparse matrix}, shape (n_samples, n_features)) – Input data samples.
- Returns:
y – Predicted outputs for inputs X.
Attention
predictalways returns a dense matrix of predicted outputs – unlike infit(), this may cause memory issues at high number of outputs and very high number of samples. Feed data by smaller batches in such case.- Return type:
ndarray, shape (n_samples,) or (n_samples, n_outputs)
- score(X, y, sample_weight=None)¶
Return the coefficient of determination of the prediction.
The coefficient of determination
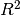 is defined as
is defined as
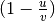 , where
, where 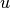 is the residual
sum of squares
is the residual
sum of squares ((y_true - y_pred)** 2).sum()and is the total sum of squares
is the total sum of squares ((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a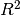 score of 0.0.
score of 0.0.- Parameters:
X (array-like of shape (n_samples, n_features)) – Test samples. For some estimators this may be a precomputed kernel matrix or a list of generic objects instead with shape
(n_samples, n_samples_fitted), wheren_samples_fittedis the number of samples used in the fitting for the estimator.y (array-like of shape (n_samples,) or (n_samples, n_outputs)) – True values for X.
sample_weight (array-like of shape (n_samples,), default=None) – Sample weights.
- Returns:
score –
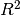 of
of self.predict(X)w.r.t. y.- Return type:
Notes
The
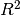 score used when calling
score used when calling scoreon a regressor usesmultioutput='uniform_average'from version 0.23 to keep consistent with default value ofr2_score(). This influences thescoremethod of all the multioutput regressors (except forMultiOutputRegressor).
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
**params (dict) – Estimator parameters.
- Returns:
self – Estimator instance.
- Return type:
estimator instance